Christine
M.
Povinelli, Ph.D.
Computational Detection of Chromosome Structure and
Function |
|
 |
Contact
Chris Povinelli:
cmpov@swbell.net
Skype: christine.povinelli
Ph.D. 1987
Georgia Institute of Technology
Molecular Biology and Genetics
|
|
OVERVIEW
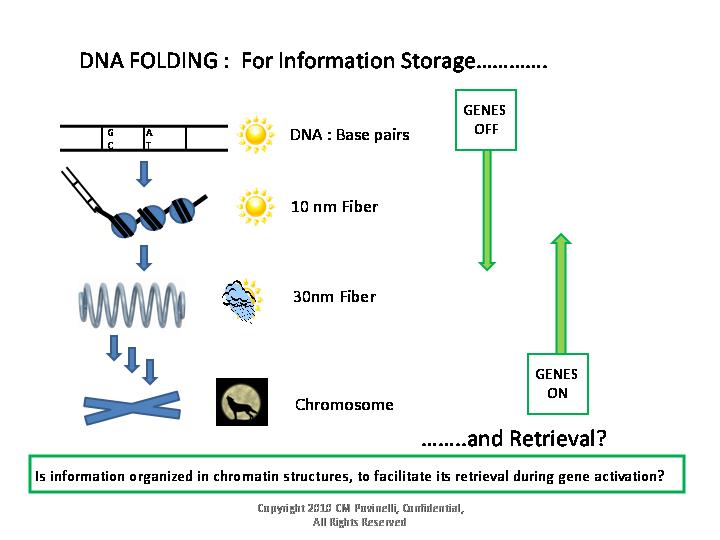
For management and protection, the large amounts of DNA in animal cells
are highly folded into compact storage structures called chromosomes.
My
research asks whether information involved in gene control is precisely
organized in the chromosomes of eukaryotes, whether this organization
describes how DNA is folded in chromosomes, and how defects in this
organization may contribute to human genetic disease.
For example, in eukaryotic 30nm
chromatin
fibers, the potential organization of multiple regulatory
elements for two different genes might be simplistically visualized as
shown in this
illustration :
In order for a gene to be expressed, the DNA containing the gene, and
the gene's regulatory regions, must be unfolded from highly condensed
chromosome structure.
In addition, several gene regulatory regions must often locate and
physically interact with each other through bound gene
regulatory proteins. These multiple regulatory regions, involved in the
control of one gene, may be
highly separated along the linear DNA.
Since the unfolding of chromatin
structure and the interaction of distant gene control regions are both
hallmarks of the early stages of gene activation, it is possible that
gene control regions are ‘pre-organized’ in relatively close proximity
in folded 30nm fibers.
As these structures begin to open and unfold, gene regulatory
proteins could, potentially, bind and interact with all of the
relatively localized regulatory regions, before the DNA is in a more
unfolded, linear, form. Detection of periodically distributed
information, with a period length equivalent to one turn of a 30nm
fiber, could therefore, describe both structural (i.e. circumference)
and functional (gene regulatory) aspects of the 30nm fiber.
|
|
|
|
 |
|
Power Point Presentation
Contains animations
Run as a slide show
Structure and Function of 30nm
Fibers Deduced From the Linear
Distribution of Genetic Information.
Part One: The Logical Framework
A brief and simplified overview of the research approaches.
|
|
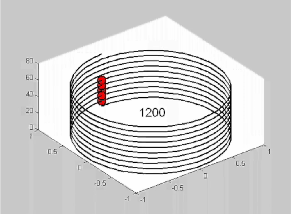
Short video made
using MATLAB:
A Graphical
Illustration of Periodicity Detection
This video shows how
information (red
dots)
that is linearly spaced, with a period of 1200BP, is distributed when
it is coiled at various circumferences. The relative locations of the
information at coil circumferences between 1325 and 575 BP are shown.
The information is vertically aligned only at circumferences of 1200 BP
(every rung) and 600 BP (every other rung). The occurrence of
vertically
aligned information is most densely aligned at the true period of
1200 BP.
Note that this type of analysis can be used both to detect the
degree to which linear information is periodically distributed and,
separately, to model the relative locations of information in 3D
solenoidal data sets. |
|
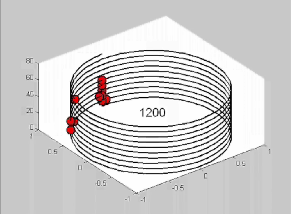
Short video made
using MATLAB:
A Graphical
Illustration of Periodicity Detection in a Split Signal with Some Noise
This short video is the same as
the one to the left but with simple
1200 BP linear periodicity imprecisely split by approximately 200 BP.
The underlying period of 1200 BP is still detected by noting the
circumference at which the red dots are most tightly, vertically,
aligned. |
|
|
|
|
|
SIGNIFICANCE
Health:
Demonstration that gene control
sequences are precisely located in
three dimensions in chromosomes would be a strong indication that this
organization is important for some process, the most likely of these is
for proper gene expression. This research is developing unique
computational tools for classifying the types of aberrations in
three-dimensional organization that can lead to genetic disease. The
insights derived from these studies can then be used to design
approaches for their correction.
Bioinformatics:
Three-dimensional analysis of human
genetic information represents an
entirely new field of study that can be referred to as‘Chromosomics’-
the study of chromosome structure and function based on the
distribution of genomic sequence data.
This is a
profound break from the current linear methods used to display and
computationally analyze genomic data. It is analogous to how protein
biologists only gain a complete understanding of protein structure and
function, when they determine how the linear string of amino acids is
folded in three dimensions. Development of computational approaches for
studying the structure of human chromosomes is necessary since
chromosome complexity and fragility has made them difficult to define
experimentally.
Computational
and Applied Mathematics: This data set, and the
methods developed during the course of this research for its characterization, represent a completely new
area of applied computational research. Prior to this research there
were no methods available for examining these types of questions.
|
|
|
|